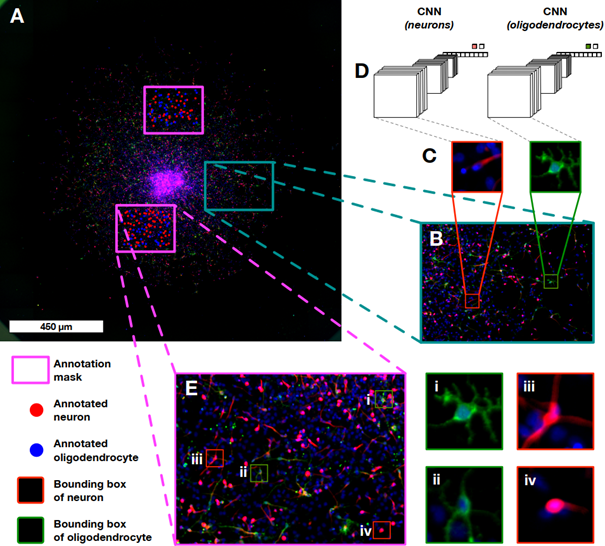
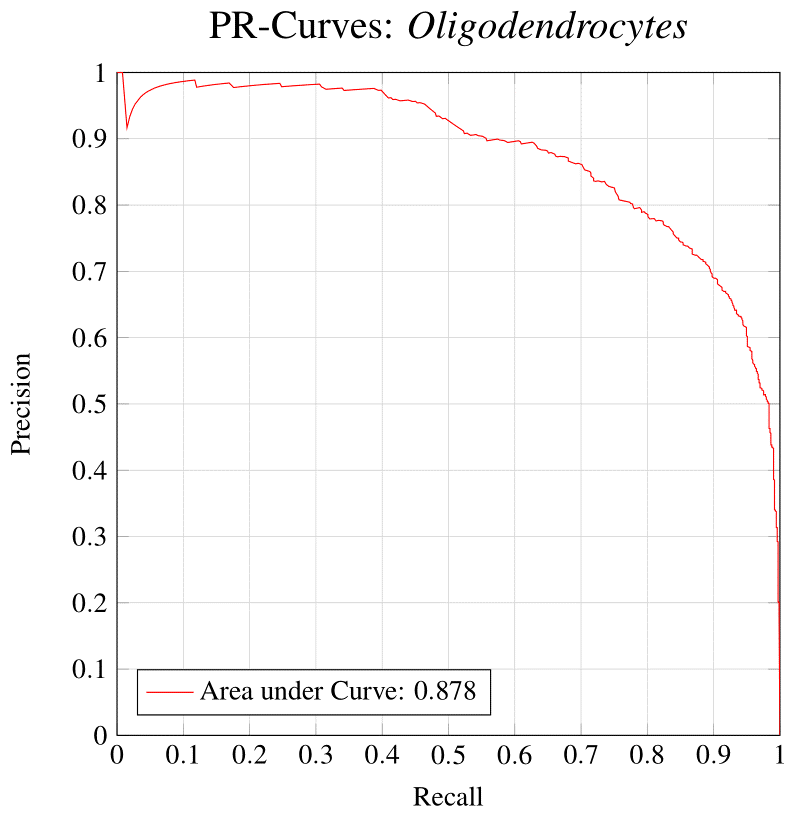
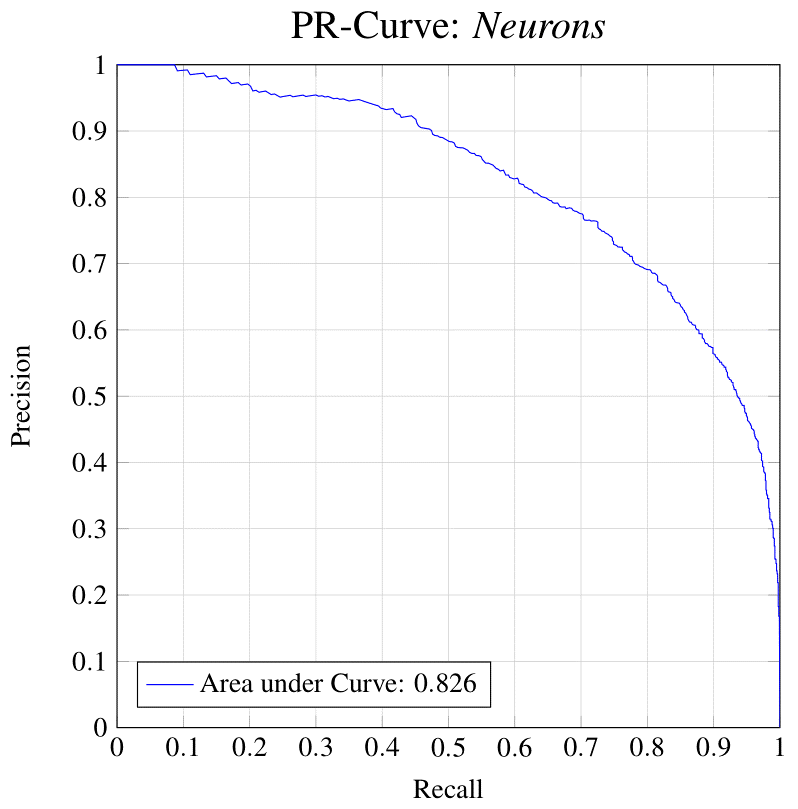
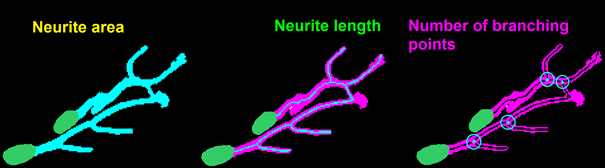
Neuronal identification/quantification
Manual
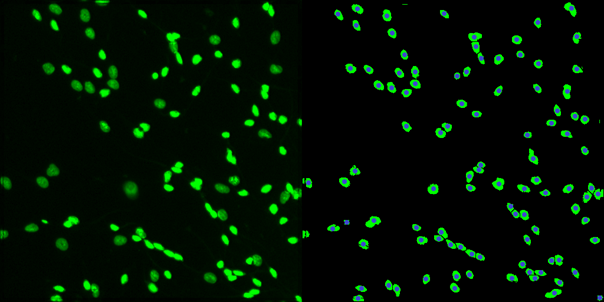
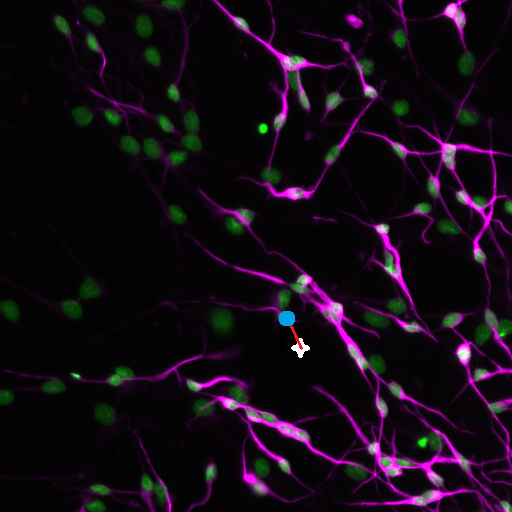
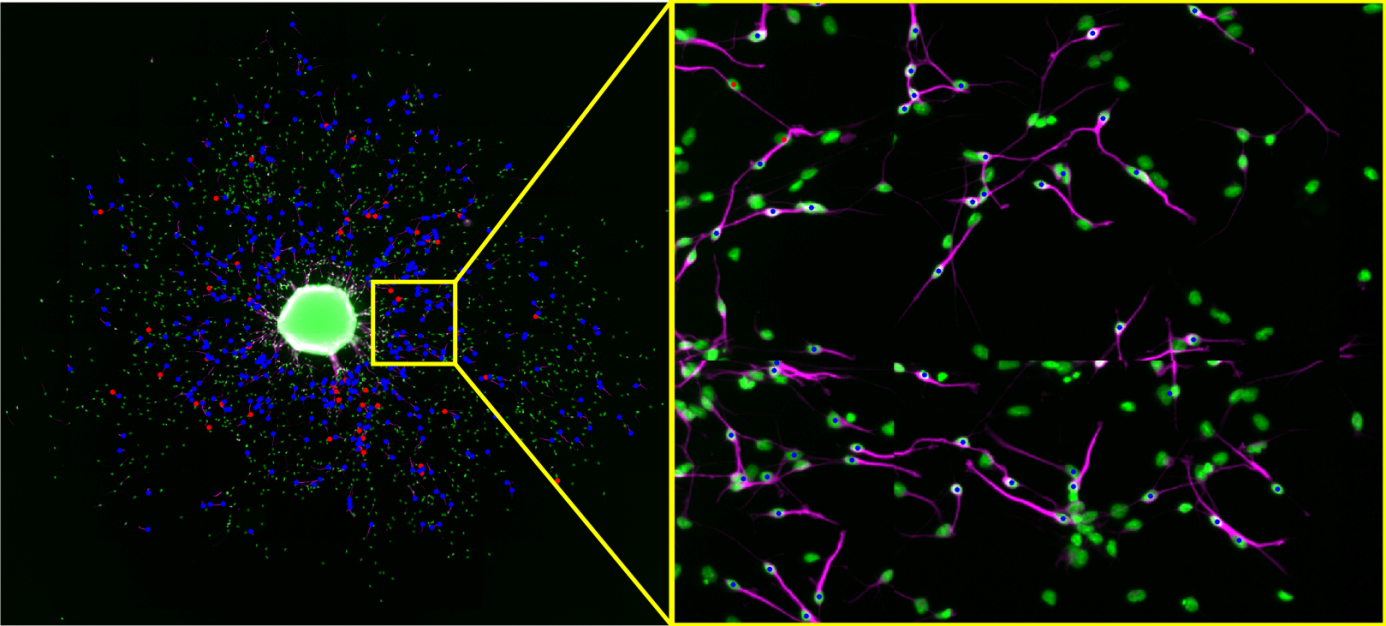
OmniSphero provides the user with a manual counting tool, to manually quantify neurons. This counting tool, saves all coordinates of the user defined neurons by mapping the clicking position to the closest surrounding nucleus.
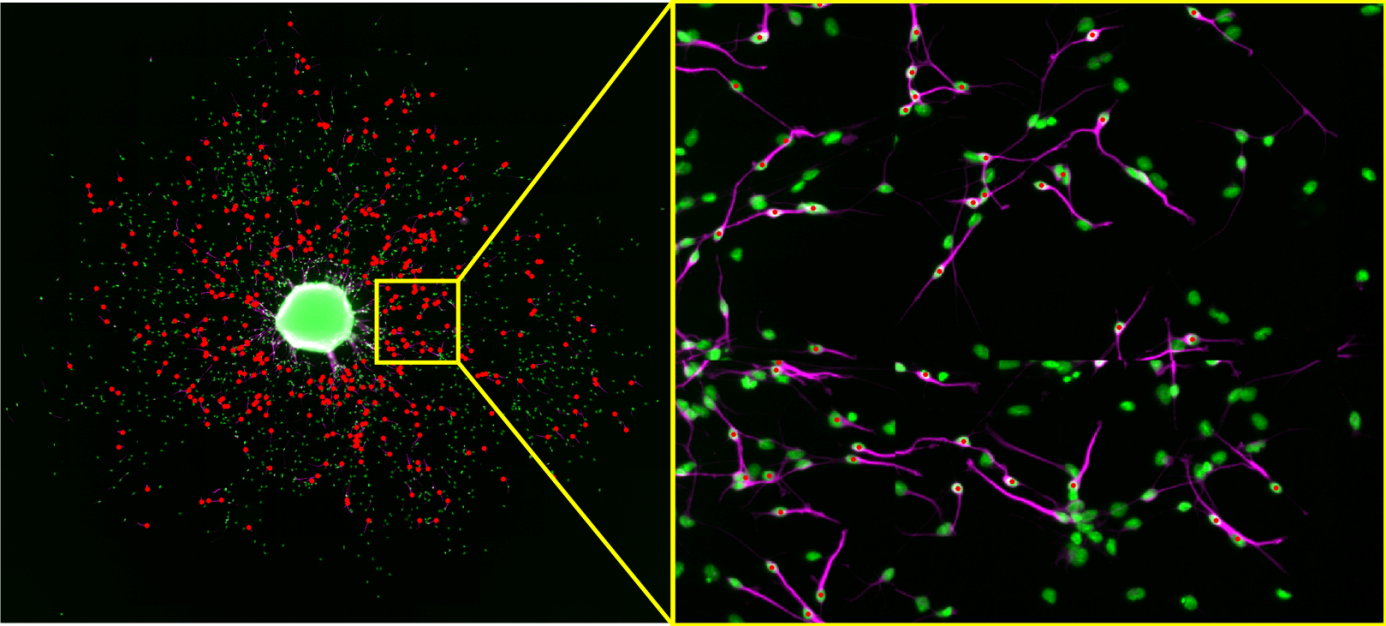
Figure 4: The user clicked position is shown as a white cross and is automatically mapped to the closest nuclei (blue dot)
Automated evaluation with classical image analysis
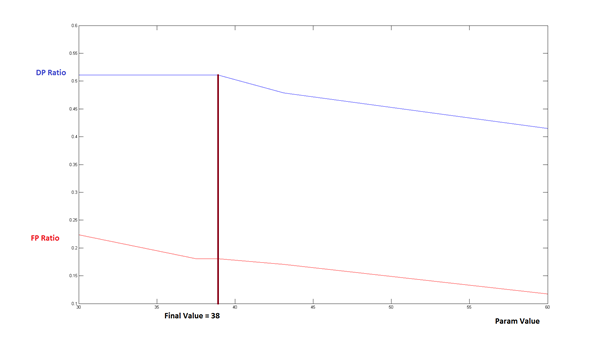
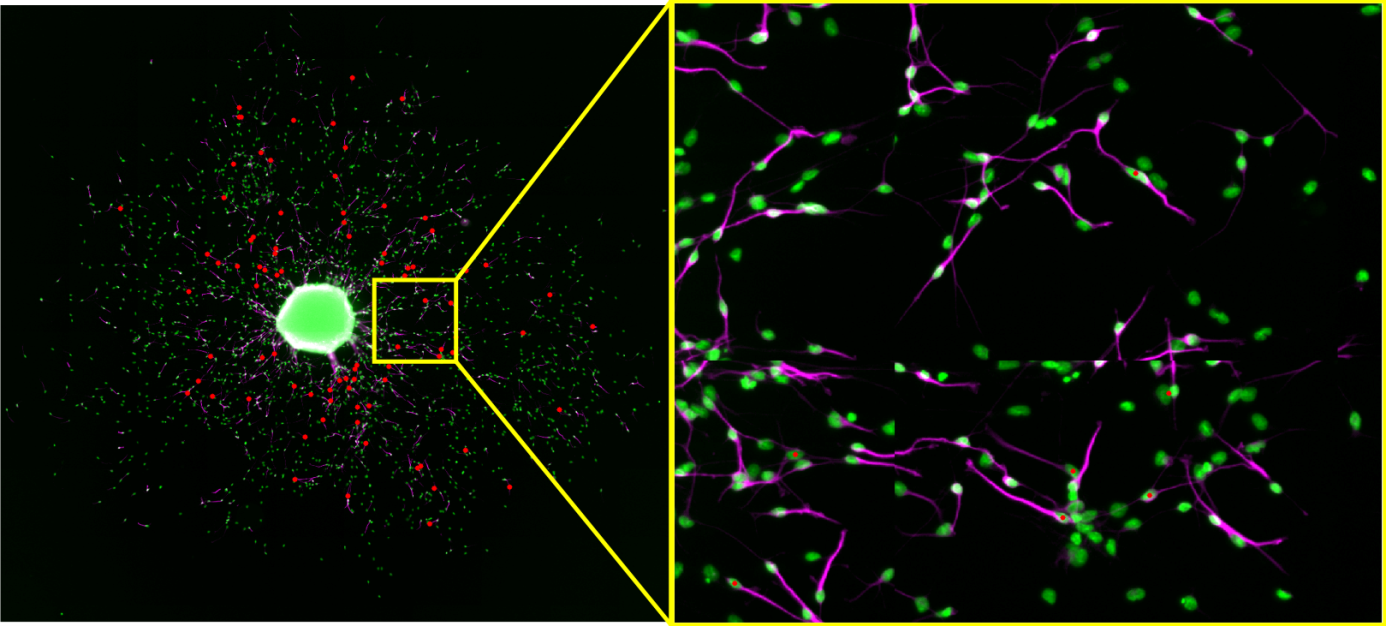
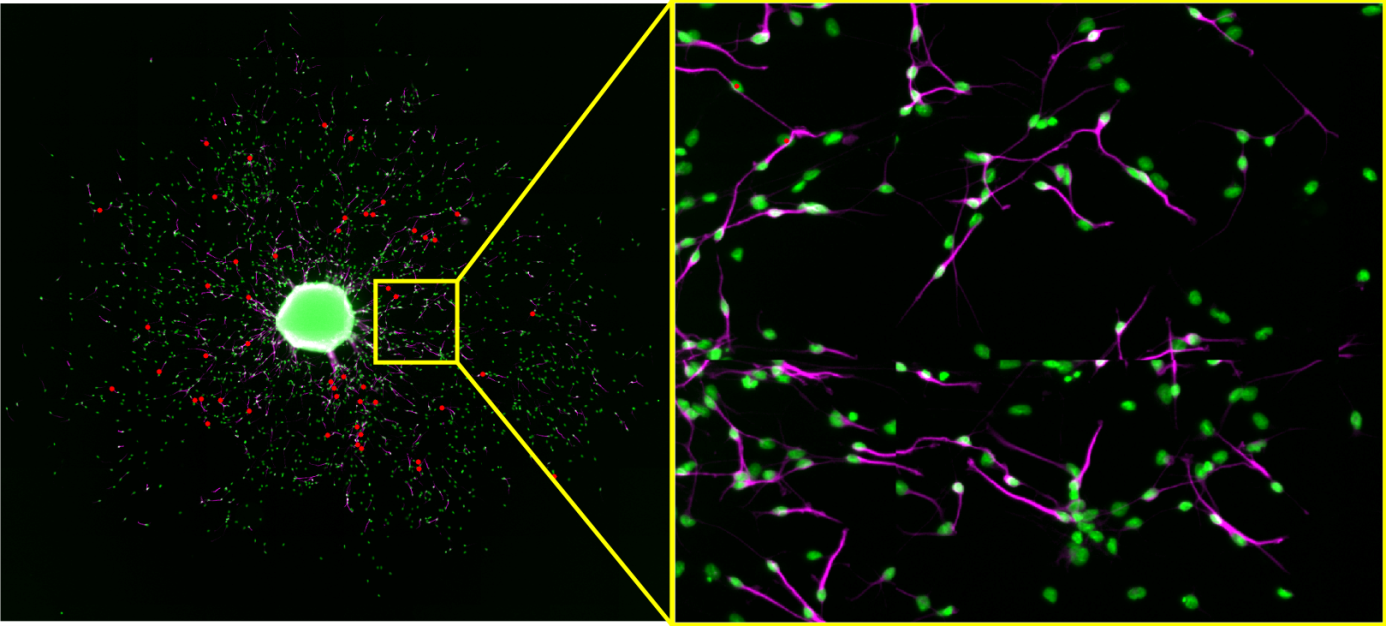
OmniSphero comprises a self-developed algorithm called ‘Neuronal Tracer’ to quantify neurons in a heterogeneous cell population with varying cell densities. Most software requires the user to adjust multiple parameters on several sample images to optimize shape recognition, which results in a time consuming optimization procedure. Therefore, we implemented a new concept in ‘OmniSphero’, in which the user defines a neuron by manual assignment rather than by defining an object over parameter settings using the manual counting tool described above. Consequently, the program optimizes automatically all parameters to fulfill a user-defined maximal false positive (FP)-rate.
Figure 5: Optimization of one parameter within in the multiparameter analysis (here minimal skeleton length in pixel). The parameter is iterated until the best ratio between true positives (Detection power (DP)) and false positives (False positive (FP)-rate) is achieved, without extending the FP-rate above the maximal user defined value.
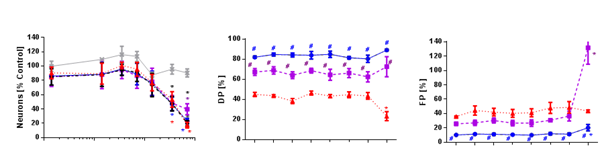
Results can be plotted as dose response curves to study the dose dependent effect of a substance on neuronal differentiation. Additionally coordinates of neurons obtained by other software can be imported into OmniSphero and DP and FP-rates can be analogous obtained by comparing them to the manually identified coordinates
Figure 6: Comparison of different automated methods in terms of accuracy and precision to identify neurons. In the left image dose response curves are shown in red blue and magenta for automated methods and in black for the manual evaluation as gold standard. Additionally the influence on viability is shown in grey. The middle and right image show the DP and FP-values for the corresponding algorithms.