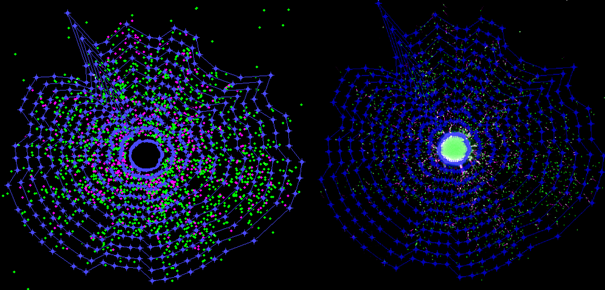
Radial migration is assessed by recognizing the sphere-core and defining it as the inner boarder of the migration area. The outer boarder is calculated by using the density of coordinates of the identified centroids of cell nuclei and the intensity information of the nucleus channel. The migration distance is subsequently calculated as the mean distance between these two boarders. Since only the raw images and the centroids of cell nuclei are required to assess this endpoint, centroids of cell nuclei obtained by different software can be imported in Omnisphero and used for the evaluation.
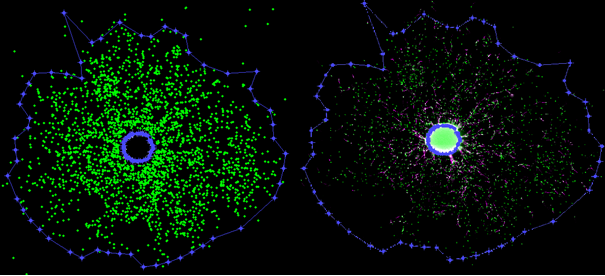
Figure 14: Assessment of the migration distance: The left image shows the point cloud of the centroids of identified nuclei. The sphere core is the brightest object within the migration area and is automatically removed by OmniSphero. The removed sphere-core defines the inner boarder of the migration area. The density of nuclei centroids is used to identify the outer boarder of the migration area. The right image shows the resulting mask of the migration area on the original image.