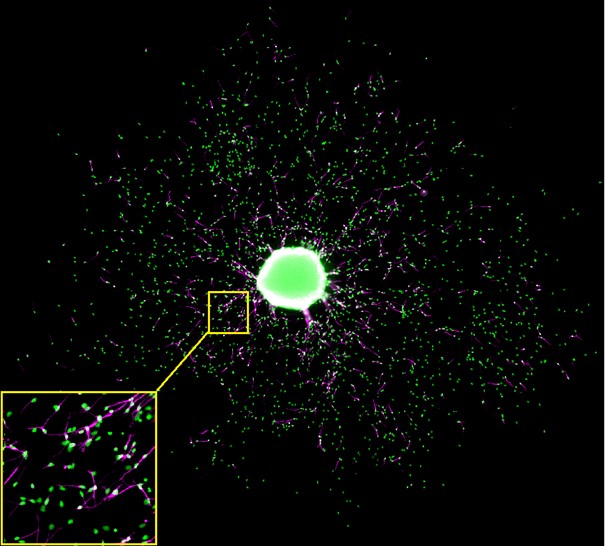
OmniSphero was developed to analyze and visualize relevant endpoints of the ‘Neurosphere Assay’ which is based on three dimensional (3D) neural progenitor cell (NPC) clusters which are able to mimic basic processes of brain development (proliferation, migration, differentiation and apoptosis) in vitro. OmniSphero comprises algorithms to analyze conventional endpoints like nucleus quantification, neuronal quantification and neuronal morphology as well as sphere-specific endpoints, which originate from characteristic features of the system, like radial glia migration distance and neuronal migration. Sphere-specific endpoints can only be analyzed on overview images of an entire well. Since a certain resolution is required to guarantee an accurate neuronal identification, the well has to be imaged in small image extracts. In our case the images were obtained with an ArrayScan VTI device (Thermo Fischer) using a 20x objective resulting in 196 images per well. OmniSphero is able to automatically compose those images to an image montage.
Image montage of a five days differentiated human neurosphere double stained for cell nuclei with Hoechst (green) and betaIII-tubuline for neurons (magenta). The yellow box shows a magnification of one of the image extracts the image montage is composed of.
Furthermore OmniSphero was especially designed to compare different automated methods, including self-developed algorithms, in their accuracy and precision of neuronal identification with the manual evaluation as a gold standard. This enables the user to identify the most appropriate software for his or her cell system. All algorithms of OmniSphero do not require any previous knowledge by the user, due to the implemented multiparameter approach which uses the users manually identified objects to assess the most adequate parameter set.